March 23, 2012
A glow coming from the glassy shell of microscopic marine algae called diatoms could someday help us detect chemicals and other substances in water samples. And the fact that this diatom can glow in response to an external substance could also help researchers develop a variety of new, diatom-inspired nanomaterials that could solve problems in sensing, catalysis and environmental remediation.
Fluorescence is the key characteristic of a new biosensor developed by researchers at the Department of Energy's Pacific Northwest National Laboratory. The biosensor, described in a paper published this week in the scientific journal PLoS ONE, includes fluorescent proteins embedded in a diatom shell that alter their glow when they are exposed to a particular substance.
"Like tiny glass sculptures, the diverse silica shells of diatoms have long intrigued scientists," said lead author and molecular biologist Kate Marshall, who works out of PNNL's Marine Sciences Laboratory in Sequim, Wash. "And the way our biosensor works could make diatoms even more attractive to scientists because it could pave the way for the development of novel, synthetic silica materials."
Diatoms are perhaps best known as the tiny algae that make up the bulk of phytoplankton, the plant base of the marine food chain that feeds the ocean's creatures. But materials scientists are fascinated by diatoms for another reason: the intricate, highly-ordered patterns that make up their microscopic shells, which are mostly made of silica. Researchers are looking at these minuscule glass cages to solve problems in a number of areas, including sensing, catalysis and environmental remediation.
PNNL Laboratory Fellow and corresponding author Guri Roesijadi found inspiration for this biosensor in previous work by other researchers, who showed it's possible to insert proteins in diatom shells through genetic engineering. Using that work as a starting point, Roesijadi, Marshall and their PNNL colleagues aimed to use fluorescent proteins to turn diatoms into a biosensor. They specifically aimed to create a reagent-less biosensor, meaning one that detects a target substance on its own and without depending on another chemical or substance.
Well-equipped diatom
As a test case, the PNNL team inserted genes for their biosensor into Thalassiosira pseudonana, a well-studied marine diatom whose shell resembles a hatbox. The new genes allowed the diatoms to produce a protein that is the biosensor.
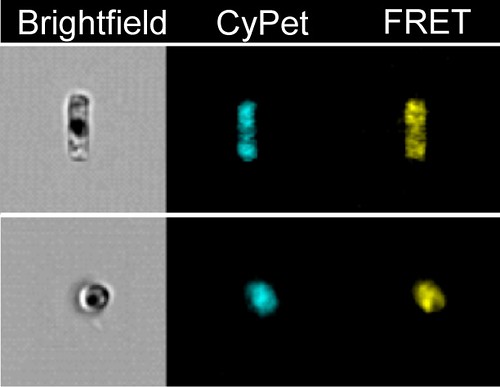
At the heart of the biosensor is the ribose-binding protein, which, as the name suggests, attaches to the sugar ribose. Each ribose-binding protein is then flanked by two other proteins — one that glows blue and another that glows yellow. This three-protein complex attaches to the silica shell while the diatom grows.
In the absence of ribose, the two fluorescent proteins sit close to one another. They're close enough that the energy in the blue protein's fluorescence is easily handed off, or transferred, to the neighboring yellow protein. This process, called fluorescence resonance energy transfer, or FRET, is akin to the blue protein shining a flashlight at the yellow protein, which then glows yellow.
But when ribose binds to the diatom, the ribose-binding protein changes its shape. This moves the blue and yellow fluorescent proteins apart in the process, and the amount of light energy that the blue protein shines on the yellow protein declines. This causes the biosensor to display more blue light.
Microscopic light show
Regardless of whether or not ribose is bound to the diatom's biosensor, the biosensor always emits some blue or yellow glow when it's exposed to energy under a microscope. But the key difference is how much of each kind of light is displayed.
The PNNL team distinguished between light from the two proteins with a fluorescence microscope that was equipped with a photon sensor. The sensor allowed them to measure the intensities of the unique wavelengths of light given off by each of the fluorescent proteins. By calculating the ratio of the two wavelengths, they could determine if the diatom biosensor was exposed to ribose, and how much of ribose was present.
The team also succeeded in making the biosensor work with the shell alone, after it was removed from the living diatom. Removing the living diatom provides researchers greater flexibility in how and where the silica biosensor can be used. The Office of Naval Research, which funded the research, believes biosensors based on modifying a diatom's silica shell may prove useful for detecting threats such as explosives in the marine environment.
"With this research, we've made our important first steps to show it's possible to genetically engineer organisms such as diatoms to create advanced materials for numerous applications," Marshall said.
Co-authors on the paper include scientists at EMSL, DOE's Environmental Molecular Sciences Laboratory at PNNL's Richland, Wash., campus. They used EMSL's mass spectrometry capabilities to verify the team had the correct ribose-binding and fluorescent proteins before adding them to the diatoms.
Source: Pacific Northwest National Laboratory (PNNL)
Additional Information:
- Kathryn E. Marshall, Errol W. Robinson, Shawna M. Hengel, Liljana Pasa-Tolic, Guritno Roesijadi, "FRET Imaging of Diatoms Expressing a Biosilica-Localized Ribose Sensor," PLoS ONE, March 21, 2012, DOI: 10.1371/journal.pone.0033771, http://dx.plos.org/10.1371/journal.pone.0033771.

0 comments:
Post a Comment